2DIR FDH
We use 2D IR to investigate active-site dynamics of Formate Dehydrogenase (FDH) via structurally guided mutagenesis of relevant active-site residues.
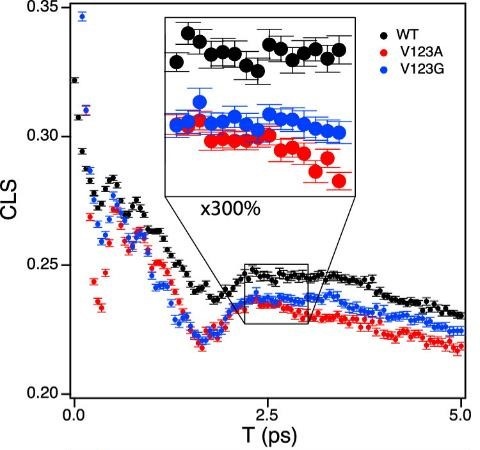
The CLS of FDH exhibits oscillations on the ps timescale
FDH CLS
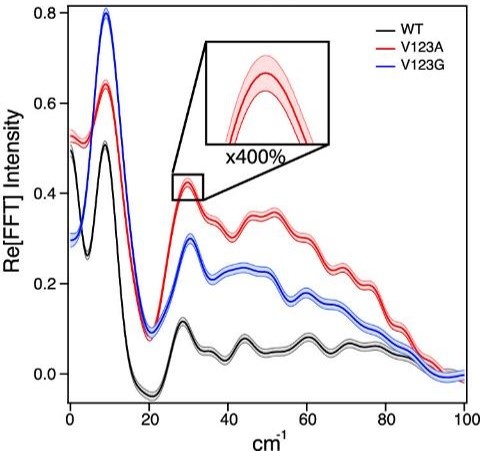
The amplitude of the oscillations increase as mutations create more space in the active site
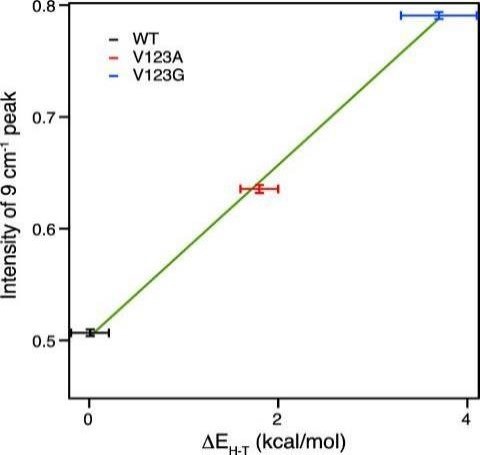
The amplitude of the frequency fluctuations correlates with donor – acceptor distance

Network residues shown on ribbon diagram of FDH
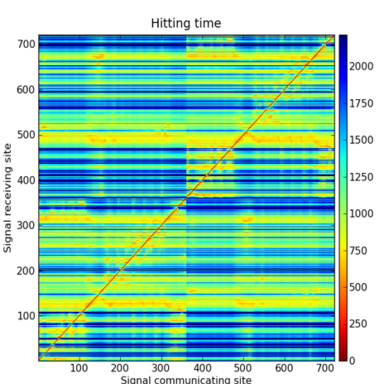
Hitting time matrix for FDH - Hitting time describes communication efficiency between residues
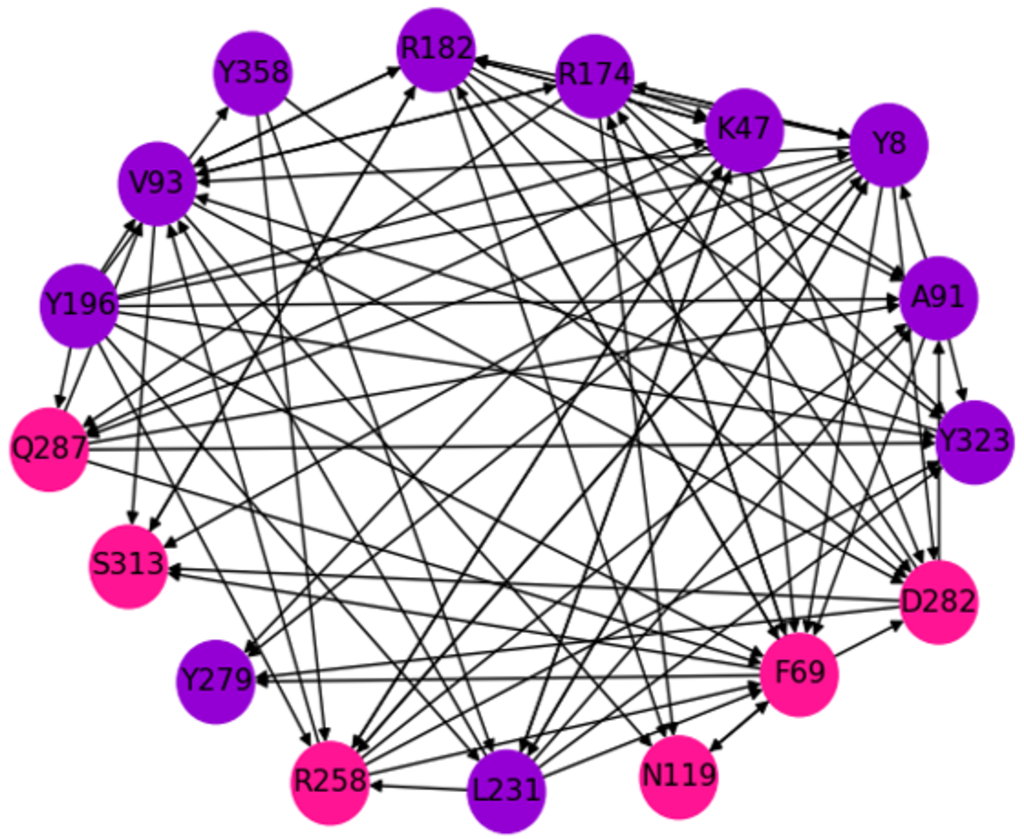
Predicted network based on hitting times using the elastic network model
We use the Elastic Network Model in our molecular dynamics simulations to interpret our 2D-IR results and to predict how perturbations affect protein motions.
Edge-Pixel Referencing
Edge-Pixel Referencing Suppresses Correlated Baseline Noise in Pump-Probe Spectra